Tutorial
Using FunVar
The Orengo group has analysed somatic, missense mutations from 22 cancer types, in order to identify mutationally enriched CATH-FunFam domain families and putative cancer driver genes (Ashford et al., 2019). FunFams are groups of functionally similar homologues in a CATH domain superfamily. For a given protein domain within a FunFam, the FunVar platform identifies mutations that are located in significantly enriched mutation clusters lying close to known or predicted functional sites in the FunFam and therefore likely to have an impact on the function. We describe these mutations as causing ‘neofunctionalisation events - NFEs’ i.e. to reflect the fact that they are modifying or changing the function.
Since FunFams are structurally coherent i.e. tend to superimpose with < 2Å RMSD, known functional sites in any of the relatives can be mapped to a structural representative for the FunFam.
The Cancer-FunVar page allows users to ‘Browse data’ on MutFams, mutations, annotations, and structural mapping of mutations as well as functional sites. A brief tutorial on how to browse the FunFam and mutations data for a particular cancer associated protein, is given below.
Example: Serine/threonine-protein kinase Chk2 (UniProt ID: O96017)
The gene CHEK2 encodes for the protein Serine/threonine-protein kinase (UniProt ID: O96017). The protein is involved in activation of DNA repair, checkpoint-mediated cell cycle arrest, and apoptosis. The example of CHEK2 will used to demonstrate CATH FunFams, functional annotations and mapping of mutation clusters in the 3D to determine whether they lie close to functional sites in the protein and could impact on function.
Step 1: Search for FunVar data using a protein query:
On the CATH-FunVar home page, click on the ‘Browse data’ tab. On the ‘Browse data’ page, click on the ‘Cancer proteins' section, in the left-hand column.
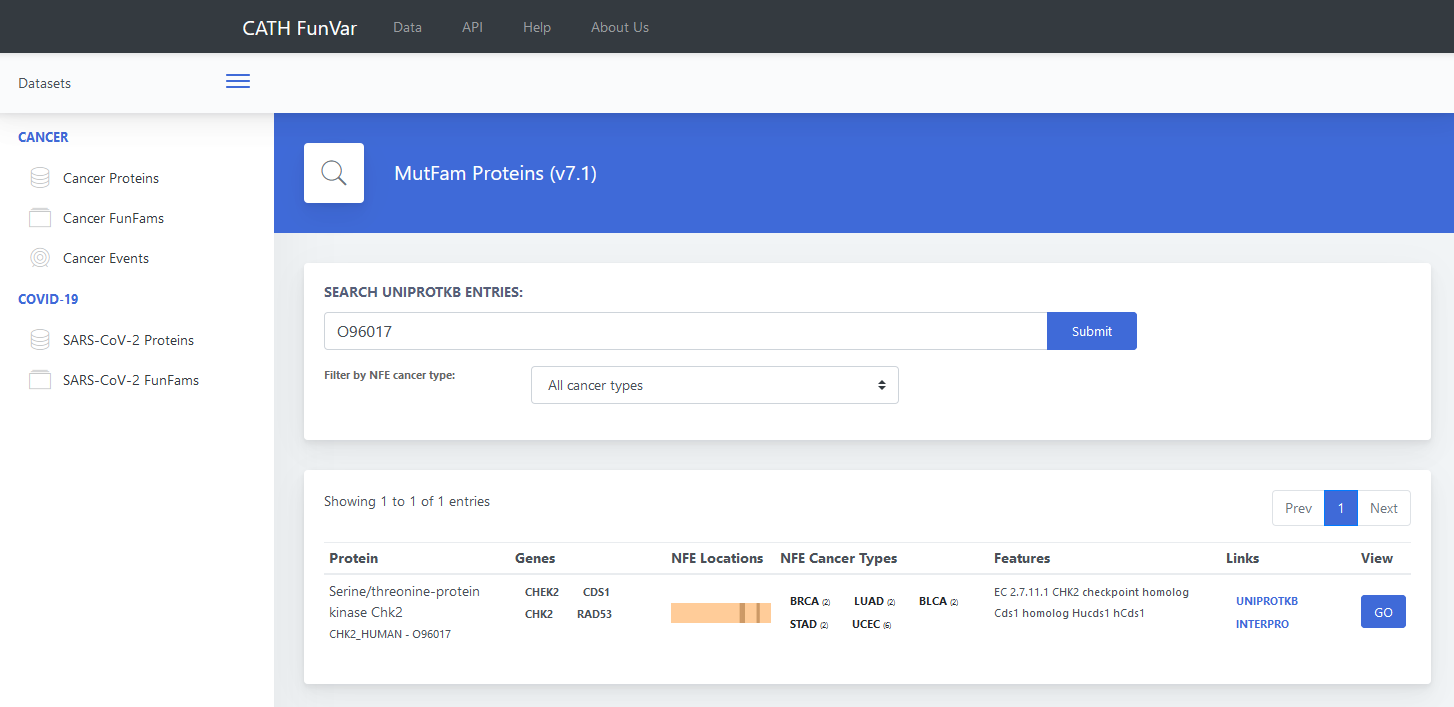
Users can search using UniProt IDs. For example, use the UniProt ID O96017, and then click on the submit button. The
result page displays the protein name, gene names (synonyms), NFE cancer types, NFE locations, features (additional
annotations such as EC number), links to the UniProtKB and InterPro page of the entry. The result page is shown in
Figure 1. The data indicates that mutations in this protein are associated with four distinct cancer types: BRCA,
LUAD, BLCA, STAD, and UCEC.
Step 2: Search for FunVar data using a protein query:
Click on the ‘Go’ button, under View column (Figure 1). This allows users to find more details
about NFEs i.e. the mutations and their location on the 3D structure, and the functional family (FunFam) to
which the protein domain associated with the entry, is assigned. The name of the FunFam reflects the most
frequently occurring GO term in the family. Details are shown in Figure 2.
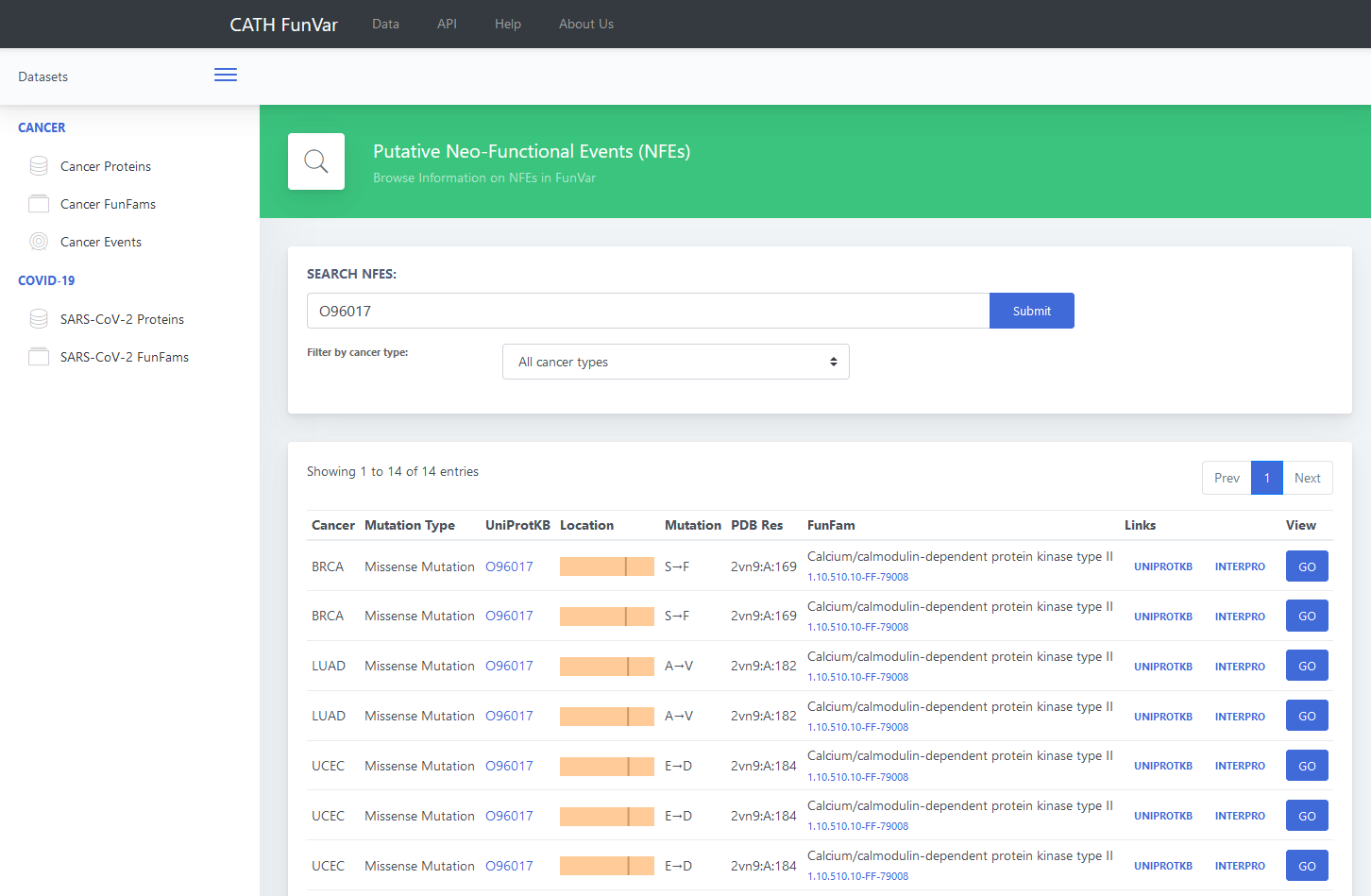
The results shown in figure 2 indicate that Serine/threonine-protein kinase Chk2 belongs to CATH-FunFam namely Calcium/calmodulin-dependent protein kinase type II (CATH FunFam ID: 1.10.510.10-ff-79008). The mutations associated with each of the four cancer types is tabulated, along with their PDB location. Users can browse structural locations of these mutations, as described in the next step.
Step 3: Browse structural mapping of functional sites, NFEs (mutations) associated with distinct cancer types
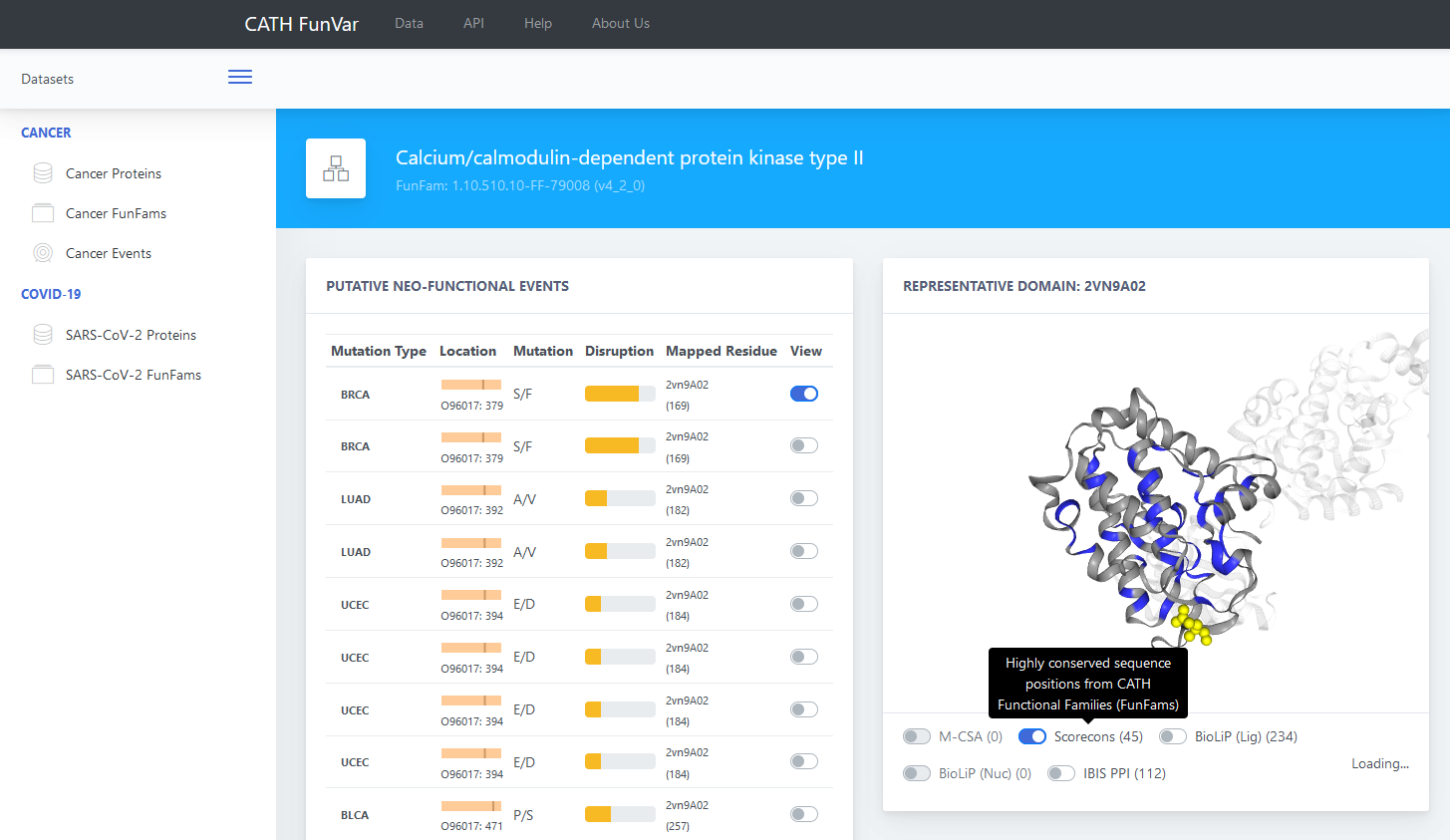
Users can browse the structural locations of NFEs/mutations by either clicking on the FunFam ID or clicking on ‘Go’ button under the View column (as shown in Figure 2). For example, the SF mutation is associated with BRCA breast cancer type. Click on either the corresponding ‘FunFam ID’ or ‘Go’ link. This provides a link to the page showing the structural mappings of each of the mutations for this entry (Figure 3). This page also shows the highly conserved, putative functional sites identified using the program Scorecons developed by the Thornton Group as well as available functional site data from M-CSA, BioLip and IBIS. Users can select particular mutations and/or functional sites and see them highlighted on the representative structure of the FunFam.
This depiction, shown in Figure 3 below, can help users to analyse the likelihood of the mutation
having an impact on the functional site and therefore the function of the protein.
We assessed the value of the mutation clusters (MutFam cluster) in CHEK2, for identifying putative drivers by their proximity to known and predicted functional sites in proteins (please refer to Ashford et al., 2019).
References
- Ashford, P., Pang, C.S.M., Moya-García, A.A. et al. A CATH domain functional family based approach to identify putative cancer driver genes and driver mutations. Sci Rep 9, 263 (2019). doi.org/10.1038/s41598-018-36401-4.
- Valdar WS. Scoring residue conservation. Proteins. 2002; 48(2):227-41. doi.org/10.1002/prot.10146